Active Motif - NOMe-Seq
Manufactured by Active Motif
NOMe-Seq for simultaneous analysis of nucleosome occupancy and DNA methylation profiles.
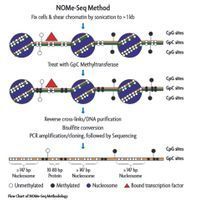
The NOMe-Seq assay works by treating fixed chromatin with a GpC methyltransferase enzyme to artificially methylate GpC dinucleotides that are not protected by nucleosomes or other proteins bound to the DNA. Due to the high abundance of GpC residues within the genome, the enzymatic treatment provides a high resolution footprint of nucleosome occupancy. Treated DNA is then subjected to bisulfite conversion and gene-specific loci are sequenced to establish the DNA methylation profile for a single DNA strand. Since GpC dinucleotides are not methylated in mammalian genomes, the endogenous CpG methylation can be distinguished from the artificially introduced GpC methylation. By combining the sequencing information for the GpC sites with the CpG sites, a profile of nucleosome occupancy and DNA methylation for a specific gene locus can be determined.
Active Questions & AnswersAsk a Question
There are no current Discussions
Need Equipment Support?
Documents & ManualsView All Documents
Features of NOMe-Seq
- Ability to simultaneously analyze nucleosome occupancy and CpG methylation on the same DNA strand * Provides a temporal relationship between nucleosome occupancy and DNA methylation * Sensitive enough to detect even subtle changes in nucleosome position * Lacks nucleosome occupancy bias that is seen with nuclease digestion techniques * Chromatin fixation also provides information regarding transcription factor binding sites
General Specifications
There are no General Specifications available.